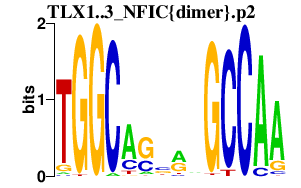
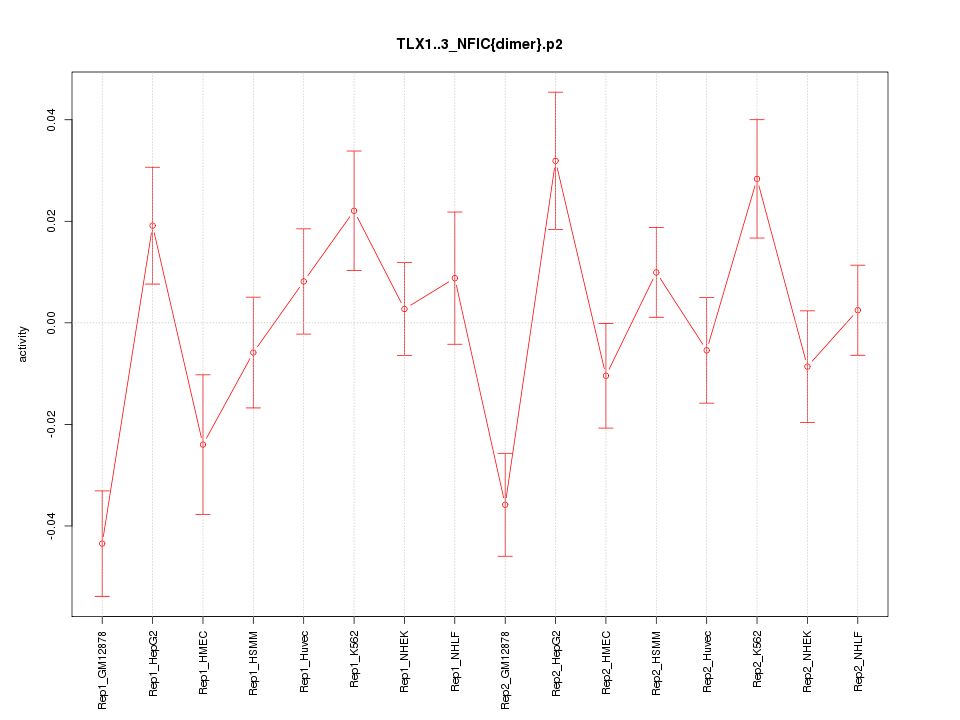
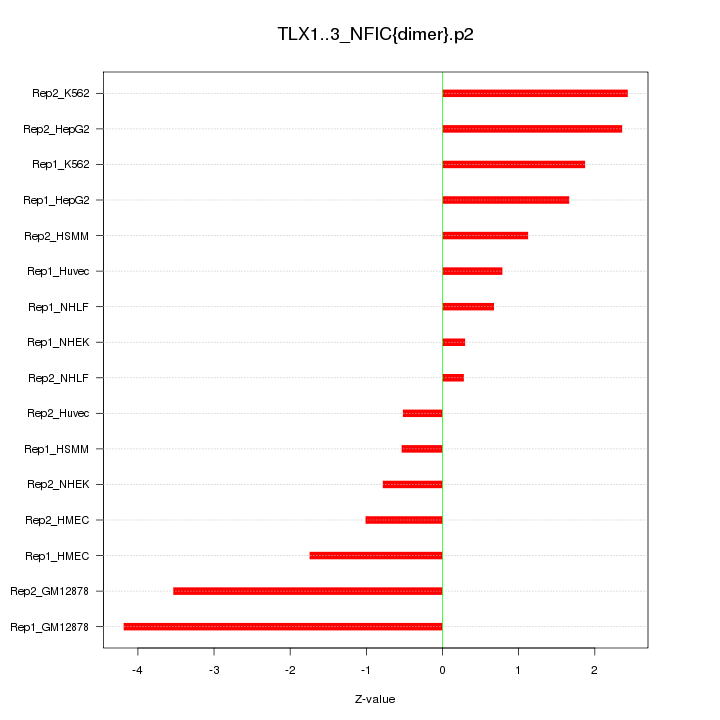
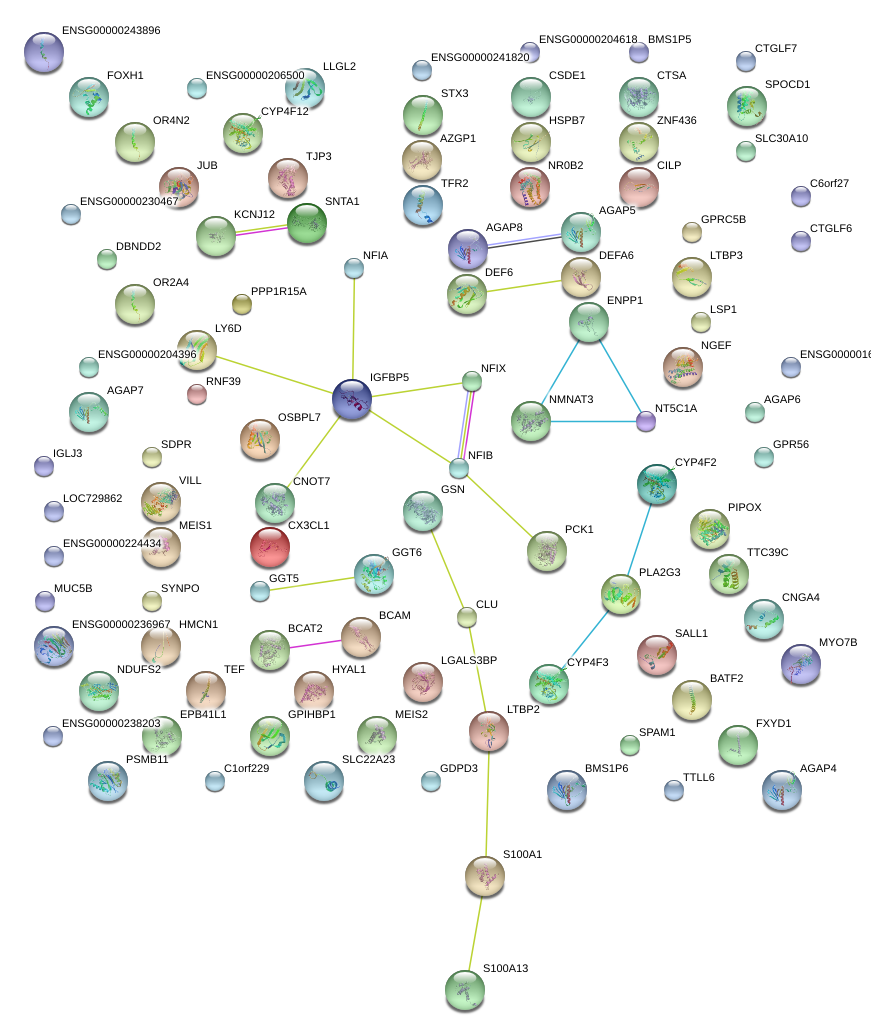
|
chr22_+_40093245
|
3.763
|
NM_001145398
|
TEF
|
thyrotrophic embryonic factor
|
|
chr1_-_16217025
|
3.076
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr11_+_1200868
|
2.662
|
NM_002458
|
MUC5B
|
mucin 5B, oligomeric mucus/gel-forming
|
|
chr17_+_24394043
|
2.168
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr22_+_21065202
|
2.166
|
|
|
|
|
chr6_-_31853027
|
2.148
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr1_+_183970093
|
2.094
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr22_+_21065134
|
2.038
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr1_-_218168548
|
1.963
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr22_+_21065207
|
1.953
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr6_-_30151526
|
1.947
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr2_-_217268492
|
1.886
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr17_-_74487527
|
1.759
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr17_+_24394281
|
1.744
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr17_-_43254117
|
1.720
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr11_+_59279481
|
1.677
|
|
STX3
|
syntaxin 3
|
|
chr19_+_15644885
|
1.644
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr1_-_218168404
|
1.642
|
|
|
|
|
chr17_-_43254132
|
1.625
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr11_-_64520964
|
1.601
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr6_+_132170847
|
1.573
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr2_-_192420168
|
1.551
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr3_+_38007264
|
1.508
|
|
VILL
|
villin-like
|
|
chr7_-_100077072
|
1.485
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr1_-_23568854
|
1.476
|
NM_001077195
|
ZNF436
|
zinc finger protein 436
|
|
chr6_-_3390197
|
1.476
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr14_+_22581215
|
1.475
|
NM_001099780
|
PSMB11
|
proteasome (prosome, macropain) subunit, beta type, 11
|
|
chr11_+_1200894
|
1.455
|
|
MUC5B
|
mucin 5B, oligomeric mucus/gel-forming
|
|
chr2_+_128009782
|
1.438
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr17_+_21249040
|
1.412
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr6_+_132170888
|
1.411
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr19_+_3659358
|
1.384
|
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr1_-_151866593
|
1.338
|
NM_001024211
|
S100A13
|
S100 calcium binding protein A13
|
|
chr22_-_22970850
|
1.314
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr7_-_143587653
|
1.306
|
NM_001005328
|
OR2A7
OR2A4
|
olfactory receptor, family 2, subfamily A, member 7
olfactory receptor, family 2, subfamily A, member 4
|
|
chr2_-_192419896
|
1.272
|
|
SDPR
|
serum deprivation response
|
|
chr8_-_145672525
|
1.263
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr7_-_99411573
|
1.220
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr1_-_39910296
|
1.194
|
NM_032526
|
NT5C1A
|
5'-nucleotidase, cytosolic IA
|
|
chr1_-_151867338
|
1.187
|
NM_005979
|
S100A13
|
S100 calcium binding protein A13
|
|
chr17_+_75507750
|
1.146
|
|
|
|
|
chr11_+_1848674
|
1.138
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr22_+_21116283
|
1.134
|
|
|
|
|
chr19_+_50004134
|
1.109
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr9_+_123083540
|
1.108
|
|
GSN
|
gelsolin
|
|
chr1_+_151867483
|
1.077
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr8_+_144366442
|
1.052
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr1_-_27113018
|
1.045
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr20_+_43468046
|
1.037
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr11_-_65082208
|
1.029
|
NM_001130144
NM_001164266
NM_021070
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr1_-_115102106
|
1.018
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr16_-_30032330
|
1.005
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr1_-_115102109
|
1.002
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr19_+_15612706
|
0.991
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr10_-_50916526
|
0.974
|
NM_001077686
|
AGAP4
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8
|
|
chr12_+_56406296
|
0.974
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr19_+_40322231
|
0.974
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr11_-_65082705
|
0.955
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr16_-_49742651
|
0.937
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr17_-_4410565
|
0.934
|
NM_001122890
NM_153338
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr16_+_56219802
|
0.918
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr22_-_29866339
|
0.917
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr2_+_66516196
|
0.917
|
|
MEIS1
|
Meis homeobox 1
|
|
chr20_+_43952997
|
0.915
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr1_-_115102124
|
0.906
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr1_-_115102084
|
0.876
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr1_-_115102138
|
0.873
|
NM_001007553
NM_007158
NM_001130523
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr22_+_21053972
|
0.833
|
|
|
|
|
chr15_-_63290839
|
0.827
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr1_-_32054164
|
0.818
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr1_+_159435728
|
0.818
|
NM_004550
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr17_+_71065478
|
0.802
|
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr17_-_44226595
|
0.775
|
NM_173623
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr20_+_55569542
|
0.768
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr14_+_19365447
|
0.768
|
NM_001004723
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2
|
|
chr14_-_22515725
|
0.765
|
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr16_+_55963899
|
0.758
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr20_-_31495086
|
0.752
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr1_-_245342150
|
0.749
|
NM_207401
|
C1orf229
|
chromosome 1 open reading frame 229
|
|
chr11_+_6216905
|
0.745
|
NM_001037329
|
CNGA4
|
cyclic nucleotide gated channel alpha 4
|
|
chr19_+_54068373
|
0.742
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr8_-_27525084
|
0.728
|
NM_001171138
|
CLU
|
clusterin
|
|
chr1_+_61319976
|
0.720
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr20_+_34206070
|
0.714
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr2_-_233586164
|
0.707
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr8_-_17148474
|
0.695
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr3_-_140879463
|
0.694
|
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr2_-_233586145
|
0.686
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr8_-_143864933
|
0.684
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr5_+_150000394
|
0.675
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr16_-_19803606
|
0.674
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr6_+_35373572
|
0.663
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr3_-_50315983
|
0.663
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr19_+_54068300
|
0.662
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr10_-_51156329
|
0.661
|
NM_001077685
|
AGAP4
AGAP7
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7
|
|
chr20_-_43953036
|
0.654
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr1_-_171286634
|
0.652
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr1_-_151866367
|
0.645
|
NM_001024212
|
S100A13
|
S100 calcium binding protein A13
|
|
chr19_-_8581559
|
0.644
|
NM_030957
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10
|
|
chr7_+_128142661
|
0.641
|
NM_032599
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr8_-_27524744
|
0.641
|
NM_203339
|
CLU
|
clusterin
|
|
chr16_-_2037819
|
0.640
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|
|
chr7_-_150283794
|
0.637
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr4_-_977163
|
0.624
|
NM_022042
NM_134425
NM_213613
|
SLC26A1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr2_+_223244606
|
0.621
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr5_-_145232723
|
0.619
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr20_-_33489383
|
0.616
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr16_-_2204776
|
0.615
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr5_+_176744037
|
0.612
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr9_-_138387903
|
0.611
|
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr12_-_27126721
|
0.605
|
NM_001080406
|
C12orf71
|
chromosome 12 open reading frame 71
|
|
chr1_-_199259133
|
0.604
|
|
KIF21B
|
kinesin family member 21B
|
|
chrX_-_54841397
|
0.603
|
NM_198510
|
ITIH5L
|
inter-alpha (globulin) inhibitor H5-like
|
|
chr1_+_54132447
|
0.601
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr11_+_119616112
|
0.598
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr17_+_58915909
|
0.594
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr2_+_185171337
|
0.591
|
NM_194250
|
ZNF804A
|
zinc finger protein 804A
|
|
chr19_-_40696393
|
0.587
|
NM_001126056
NM_001126057
NM_001126058
NM_001190347
NM_001190348
NM_001190349
NM_033317
|
DMKN
|
dermokine
|
|
chrX_-_110541029
|
0.586
|
NM_000555
|
DCX
|
doublecortin
|
|
chr2_+_24869753
|
0.584
|
NM_024322
|
CENPO
|
centromere protein O
|
|
chr6_-_638010
|
0.583
|
NM_018303
|
EXOC2
|
exocyst complex component 2
|
|
chr3_+_8750485
|
0.582
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr10_-_45662912
|
0.581
|
NM_133446
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4
|
|
chr17_-_39700960
|
0.579
|
NM_000342
|
SLC4A1
|
solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group)
|
|
chr15_-_87840780
|
0.578
|
NM_016321
|
RHCG
|
Rh family, C glycoprotein
|
|
chr8_-_17148637
|
0.575
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr2_+_66516020
|
0.563
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr10_-_69267774
|
0.561
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr3_+_9766994
|
0.561
|
|
OGG1
|
8-oxoguanine DNA glycosylase
|
|
chr14_+_94117545
|
0.551
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr3_+_9766901
|
0.550
|
|
OGG1
|
8-oxoguanine DNA glycosylase
|
|
chr6_+_34312979
|
0.549
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_38778860
|
0.548
|
|
GLO1
|
glyoxalase I
|
|
chr2_+_218896219
|
0.546
|
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr9_-_8723893
|
0.545
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_42507180
|
0.538
|
|
|
|
|
chr2_-_74588293
|
0.533
|
NM_032673
|
PCGF1
|
polycomb group ring finger 1
|
|
chr1_+_16247870
|
0.532
|
NM_001165945
|
CLCNKB
|
chloride channel Kb
|
|
chr5_+_176726918
|
0.531
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr2_-_24869718
|
0.531
|
NM_001013663
|
C2orf79
|
chromosome 2 open reading frame 79
|
|
chr10_+_94823636
|
0.530
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr12_-_120781002
|
0.529
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr6_-_43703024
|
0.527
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr1_+_199259664
|
0.526
|
|
|
|
|
chr11_+_74548472
|
0.524
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr1_-_199259447
|
0.523
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr14_+_20595820
|
0.520
|
NM_138331
|
RNASE8
|
ribonuclease, RNase A family, 8
|
|
chr11_-_118716736
|
0.513
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr10_+_685887
|
0.508
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr11_-_62213577
|
0.506
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr7_+_115952325
|
0.504
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr9_-_123301856
|
0.502
|
|
GGTA1
|
glycoprotein, alpha-galactosyltransferase 1 pseudogene
|
|
chr11_+_116205830
|
0.502
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr10_-_126097444
|
0.501
|
|
OAT
|
ornithine aminotransferase
|
|
chr3_+_9766602
|
0.500
|
NM_002542
NM_016819
NM_016820
NM_016821
NM_016826
NM_016827
NM_016828
NM_016829
|
OGG1
|
8-oxoguanine DNA glycosylase
|
|
chr9_+_130056513
|
0.497
|
|
|
|
|
chr20_-_33580861
|
0.496
|
NM_001145350
|
C20orf173
|
chromosome 20 open reading frame 173
|
|
chr3_-_31997738
|
0.492
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr22_+_20252015
|
0.489
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr2_-_74588006
|
0.488
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr7_+_6760264
|
0.483
|
NM_173565
NM_001099697
|
RSPH10B
RSPH10B2
|
radial spoke head 10 homolog B (Chlamydomonas)
radial spoke head 10 homolog B2 (Chlamydomonas)
|
|
chr2_+_97696462
|
0.480
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr8_-_145097031
|
0.479
|
NM_201380
|
PLEC
|
plectin
|
|
chr7_+_115952279
|
0.477
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr10_-_126097483
|
0.475
|
NM_000274
NM_001171814
|
OAT
|
ornithine aminotransferase
|
|
chr6_+_32057779
|
0.474
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr1_+_54786394
|
0.473
|
NM_015547
NM_147161
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr1_-_32940850
|
0.473
|
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr7_-_36991189
|
0.472
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_-_159459814
|
0.470
|
|
APOA2
|
apolipoprotein A-II
|
|
chr1_-_23683214
|
0.469
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr5_-_36277441
|
0.468
|
|
C5orf33
|
chromosome 5 open reading frame 33
|
|
chr22_-_37970902
|
0.467
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chr1_-_159435440
|
0.464
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr1_-_159459999
|
0.462
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr3_-_53855443
|
0.457
|
NM_018397
|
CHDH
|
choline dehydrogenase
|
|
chr20_-_60375695
|
0.456
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr6_-_38778893
|
0.455
|
|
GLO1
|
glyoxalase I
|
|
chr22_+_19239221
|
0.452
|
|
MED15
|
mediator complex subunit 15
|
|
chr2_+_185171537
|
0.451
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr6_-_43703057
|
0.450
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr3_-_58538110
|
0.448
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr14_+_23675233
|
0.444
|
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr17_-_71995401
|
0.441
|
NM_001005498
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr1_+_23994914
|
0.439
|
|
|
|
|
chr3_+_63928459
|
0.438
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr9_-_137731139
|
0.438
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr9_+_135389698
|
0.437
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr6_-_38778882
|
0.436
|
|
GLO1
|
glyoxalase I
|
|
chr22_+_21870293
|
0.435
|
|
|
|
|
chr5_-_36277656
|
0.433
|
NM_001085411
|
C5orf33
|
chromosome 5 open reading frame 33
|
|
chr5_+_176449156
|
0.432
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr16_+_65783571
|
0.432
|
|
E2F4
|
E2F transcription factor 4, p107/p130-binding
|
|
chr7_+_50318801
|
0.431
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr9_+_130719034
|
0.429
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr6_-_38778898
|
0.428
|
|
GLO1
|
glyoxalase I
|
|
chr11_-_85074848
|
0.426
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr2_-_218551786
|
0.423
|
|
TNS1
|
tensin 1
|
|
chr1_-_229181203
|
0.422
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr10_-_49993425
|
0.422
|
NM_001031746
NM_144984
|
C10orf72
|
chromosome 10 open reading frame 72
|